| Links |
Follow @lncrnalab |
 | Kanduri Lab Long non-coding RNA research |
| Links |
Follow @lncrnalab |
 | Kanduri Lab Long non-coding RNA research |
 Long non-coding RNAs: Overview |
 Cell cycle LncRNAs in cancer |
 LncRNAs as biomarkers in Neuroblastoma |
 Chromatin associated LncRNAs |
|
While it is relatively easy to identify transcribed lncRNAs, it remains a major challenge to characterize their functions. Thus there is a need to identify biologically relevant lncRNA with functional roles in cell differentiation, development and disease. To address this, we use novel high-throughput screening approaches to identify functional lncRNAs involved in tumor development and progression. We are on the verge of characterizing their molecular functions and demonstrate the functional relevance of these lncRNAs in tumor development and progression using different cell lines and animal model systems. We also expect to identify lncRNAs that can be used as biomarkers for human disease and as novel targets for future drug development. The following are the functional screens that we are currently using to identify biologically relevant functional lncRNAs. |
1. Cell cycle based screening of cancer associated lncRNAs | |
|
Chronic cell proliferation is considered one of the important features of cancer. Cell division cycle plays a critical role in the regulation of normal and chronic cell proliferation by responding to cell cycle regulators (cyclins, cyclin dependent kinases (CDKs), growth factors, growth suppressors, tumor suppressors and oncogenes). Cell division primarily comprises two phases: the DNA synthesis (S) phase and the mitotic (M) phase. The pathways that control DNA synthesis or replication during S-phase are critical for normal cell proliferation, as errors during DNA synthesis could result in abnormal cell proliferation. Importantly, replication occurs during different stages of S-phase. For example, replication of active chromatin regions occurs during S-phase whereas that of heterochromatin or repressive chromatin regions occur during late S-phase. Thus, characterization of mechanisms that control S-phase progression would allow us understanding the role of S-phase in normal and disease conditions. In comparison to protein-coding genes, the role of lncRNAs in S-phase regulation is still poorly understood. |
|
|
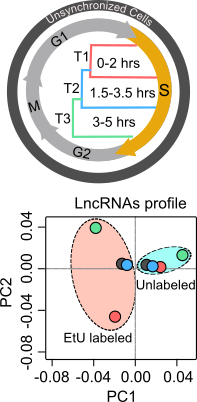
To explore the functional role of lncRNAs in S-phase progression, we need to produce time resolved transcription maps covering early, mid and late stages S-phase. To this end, we have adapted a real-time strategy "Nascent RNA Capture assay", whereby we can capture the ongoing transcription of lncRNA with high turnover in different compartments of S phase. The EtU labeled RNA along with unlabeled total RNA, representing steady state levels, collected at early, mid late stages of S phase, are subjected to deep sequencing. By applying state of the art computational tools on Etu labelled and unlabelled steady state RNA levels from early (T1), mid (T2) and late (T3) S-phase stages and unsynchronized HeLa cells, we identified 1,145 S-phase-specific temporally expressed lncRNAs. 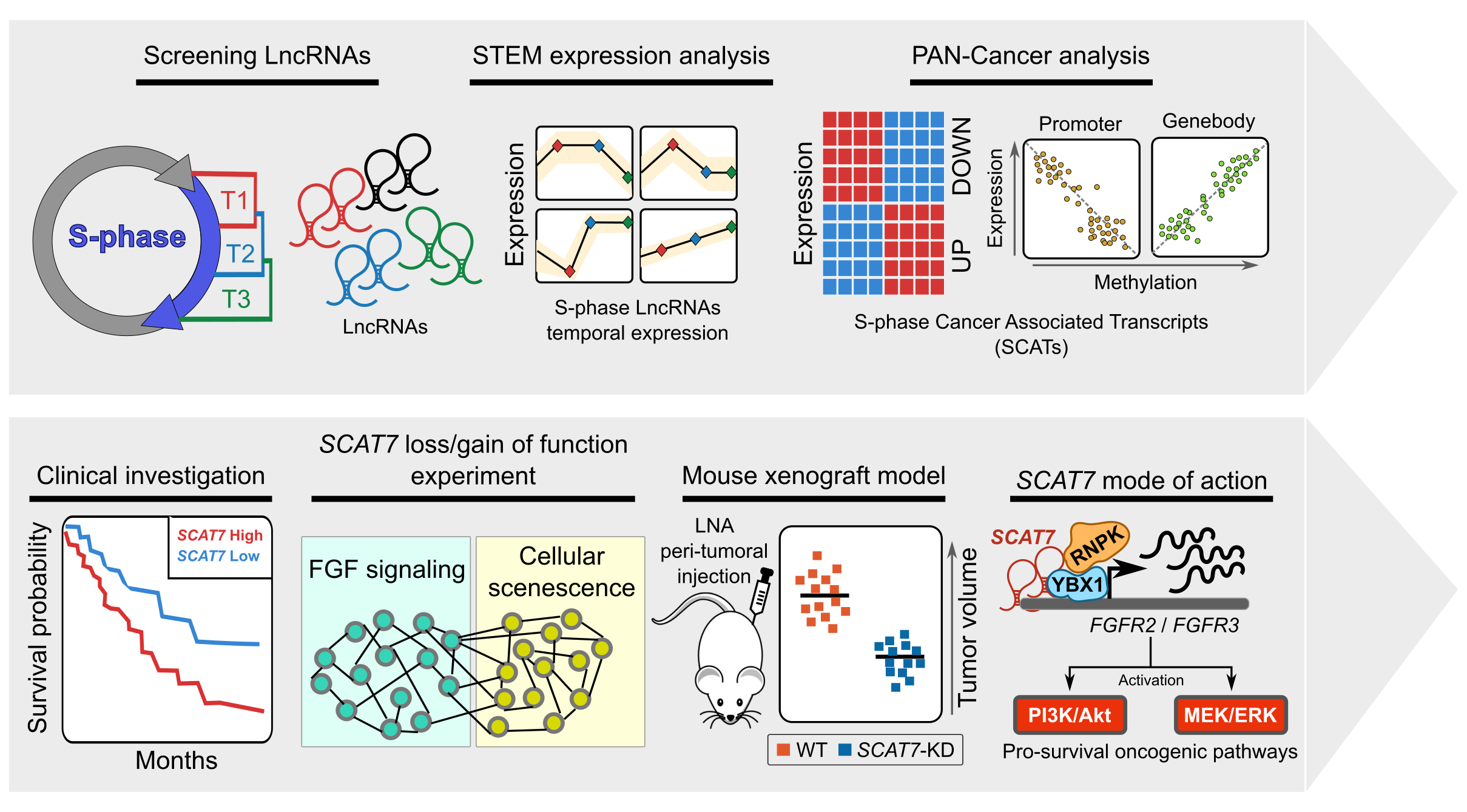 |
|
By analysing their expression levels in 6,419 solid tumors and 701 normal tissue samples from The Cancer Genome Atlas database covering 16 cancer types, we identified 570 differentially expressed lncRNAs at least in one cancer type. Likewise, our systematic and detailed clinical investigation using 14 cancer types, identified 633 independent prognostic markers. These S-phase-specific cancer-associated lncRNAs (SCATs) would serve as huge resource to explore their functional role in tumor initiation and development. Our detailed mechanistic investigations on one of the SCATs SCAT7 revealed that it is oncogenic lncRNA, which interferes with all well characterized cancer cell hall marks. 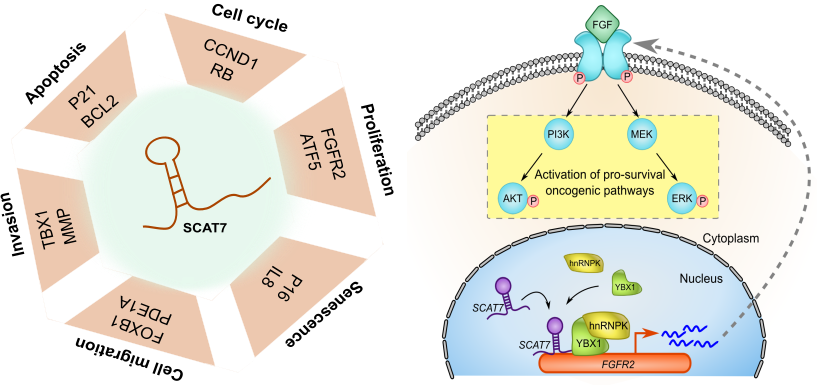
|
|
2. LncRNAs as biomarker in neuroblastoma | |
|
CASC15: By analysing the RNA-sequencing data of high- and low-risk neuroblastoma tumors we identified several differentially expressed lncRNAs. Some of the top differentially expressed lncRNA genes (NBAT1, CASC15 and its isoforms CASC15-003 and CASC15-004) map to the neuroblastoma risk-associated locus 6p22.3. Several single nucleotide polymorphisms located in the 6p22.3 locus were implicated in the metastatic high risk neuroblastoma by genome wide association studies. However, lack of any known protein coding gene in this locus made it challenging to functionally characterize the locus. We found that both NBAT1 and CASC15 (CASC15-003 and CASC15-004) are pro-neruogenic lncRNAs with high expression in Neural Crest (NC) cells and their deficiency blocks differentiation of both NC and neuroblastoma cells. 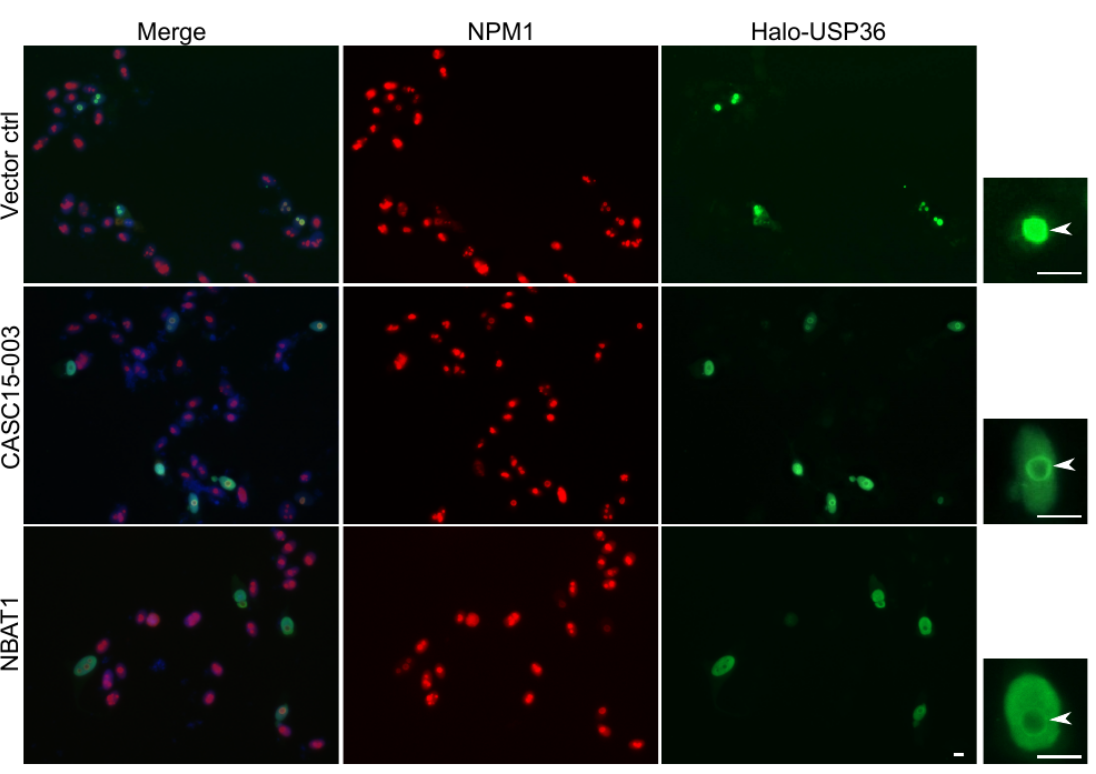 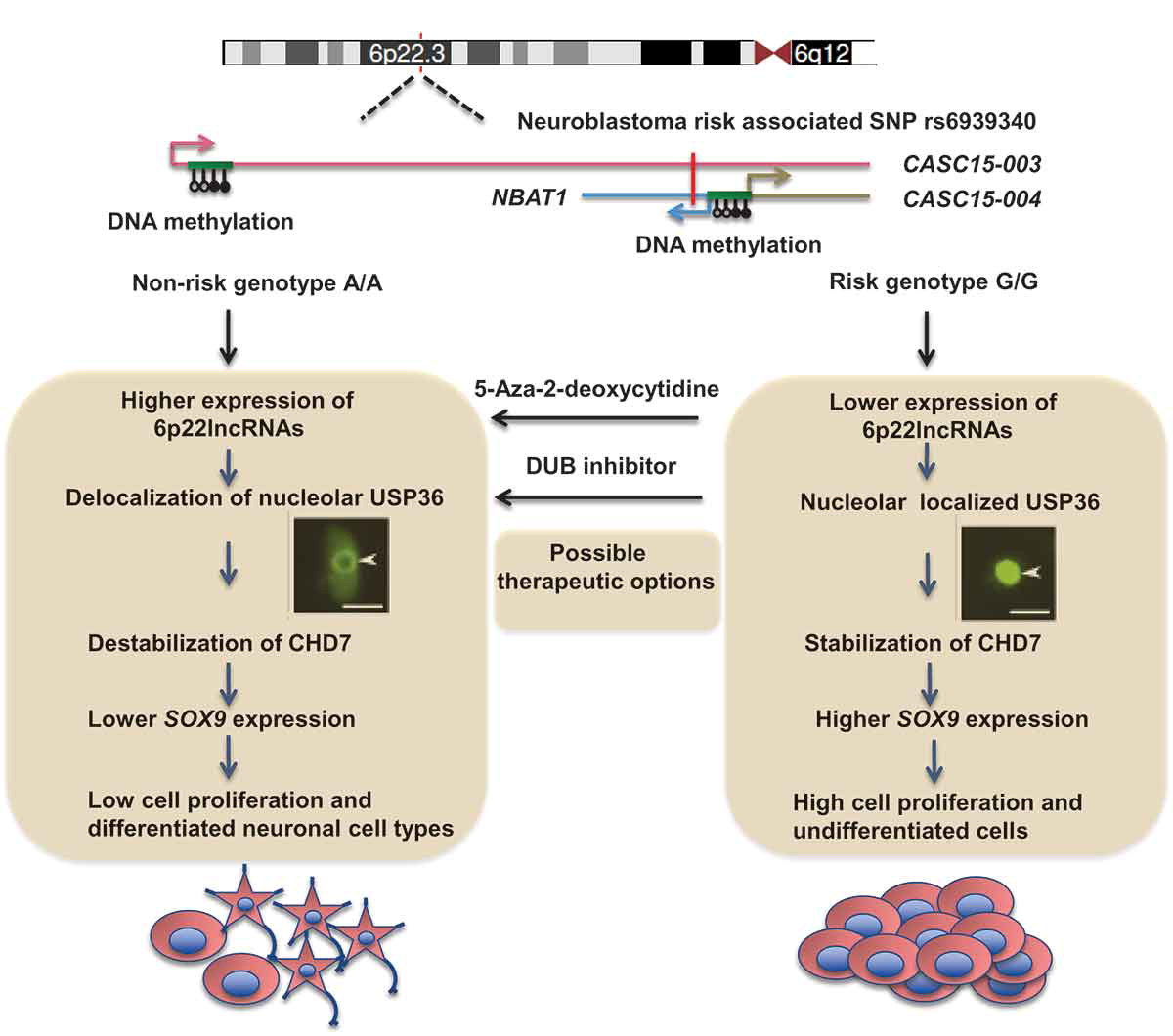 Consistent with their role in neuronal differentiation lower expression of 6p22.3 lncRNAs correlates with poor prognosis of neuroblastoma patients. Based on our published evidence, NBAT1 and CASC15-003 share similar mechanisms that underlie tumor suppression. We have shown that NBAT1 and CASC15-003 interact with a deubiquitinase protein USP36 and regulate the nucleolar localization of USP36, thereby interferes with the proteasome mediated degradation of chromatin remodeller CHD7. During differentiation of neuroblastoma cells spatial distribution of USP36 is altered: nucleolar localized USP36 gets dispersed in the nucleus but in the neuroblastoma cells lacking NBAT1 and CASC15-003, USP36 remains in the nucleolus. Nucleolar USP36 promotes CHD7 stabilization which activates the expression of pro-tumorogenic genes including SOX9. 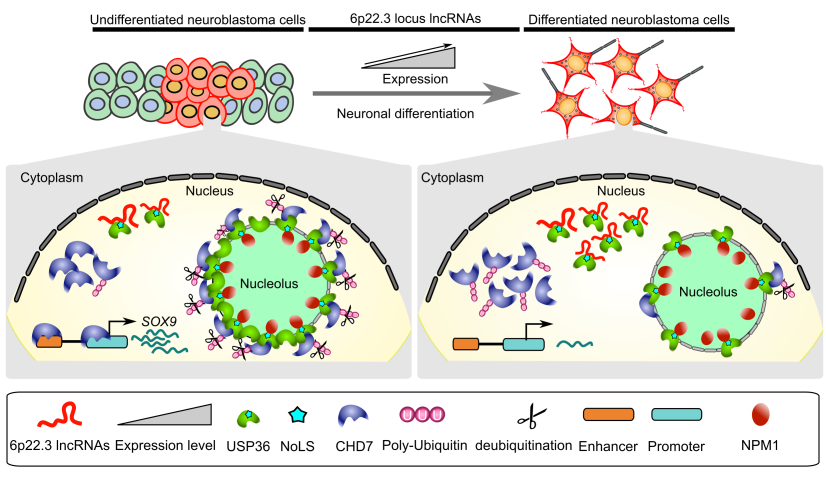
[1] Mitra S, et al. Subcellular distribution of p53 by the p53-responsive lncRNA NBAT1 determines chemotherapeutic response in neuroblastoma. Cancer researech 2020. | |
|
| |
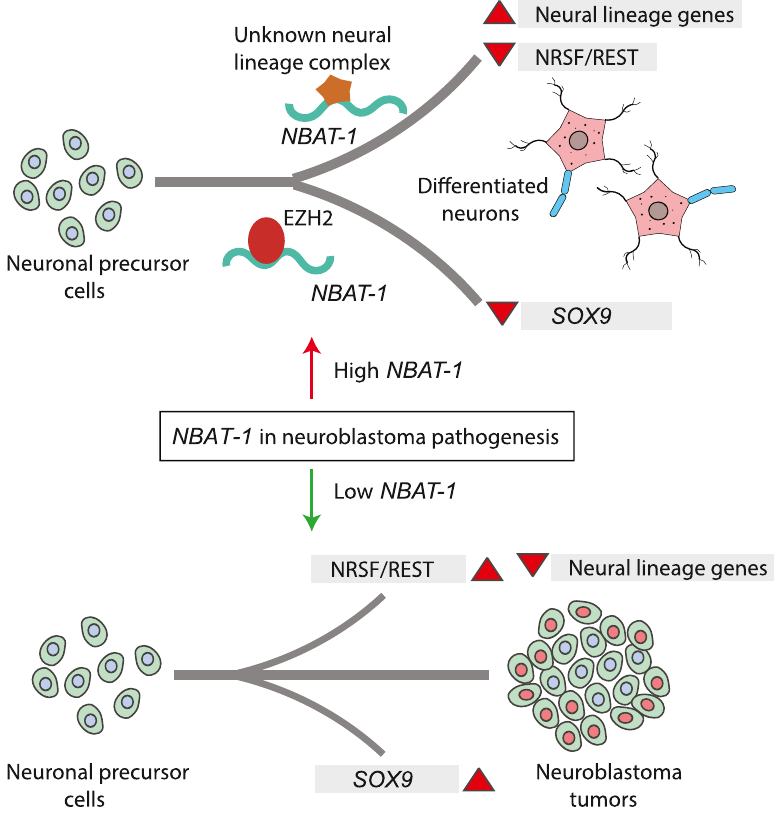 |
Model explaining the tumor suppressor and neuronal differentiation properties of NBAT-1. NBAT-1 functional interaction with EZH2, a member of PRC2 complex, controls tumor progression by suppressing protumor genes such as SOX9, VCAN, and OSMR via regulating chromatin structure. On the other hand, NBAT-1 promotes neuronal lineage commitment by suppressing NRSF/REST by interacting with an unknown neuronal-linage-specific transcriptional repressor. The NBAT-1/EZH2 interaction has no functional role in the repression of NRSF/REST. |
[4] Pandey GK, et al. Review: Long noncoding RNAs and neuroblastoma. Oncotarget 2015.
[5] Pandey GK, et al. The risk-associated long noncoding RNA NBAT-1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer Cell 2014;26;722-737.
3. Genome-wide screening of functional long noncoding RNAs | ||||
|
Chromatin associated lncRNAs (CARs): Using chromatin as a substrate lncRNAs have been shown to mediate epigenetic modifications for transcriptional regulation of gene expression. LncRNAs have the ability to both interact with and recruit chromatin modifying enzymes to catalyse the epigenetic modifications. Over the last few years, we have been trying to characterize RNAs that are part of chromatin, in particular we are interested in understanding how they are targeted to chromatin and what kind of functions chromatin bound RNAs have in gene regulation. We had previously used sucrose gradient purified chromatin to isolate chromatin bound lncRNAs and subsequently we have standardized a technique called chromatin RNA immunoprecipitation (ChRIP) to characterize the lncRNAs associated with inactive chromatin compartment as defined by distinct histone modifications.
| ||||
|
| ||||
|
|
||||
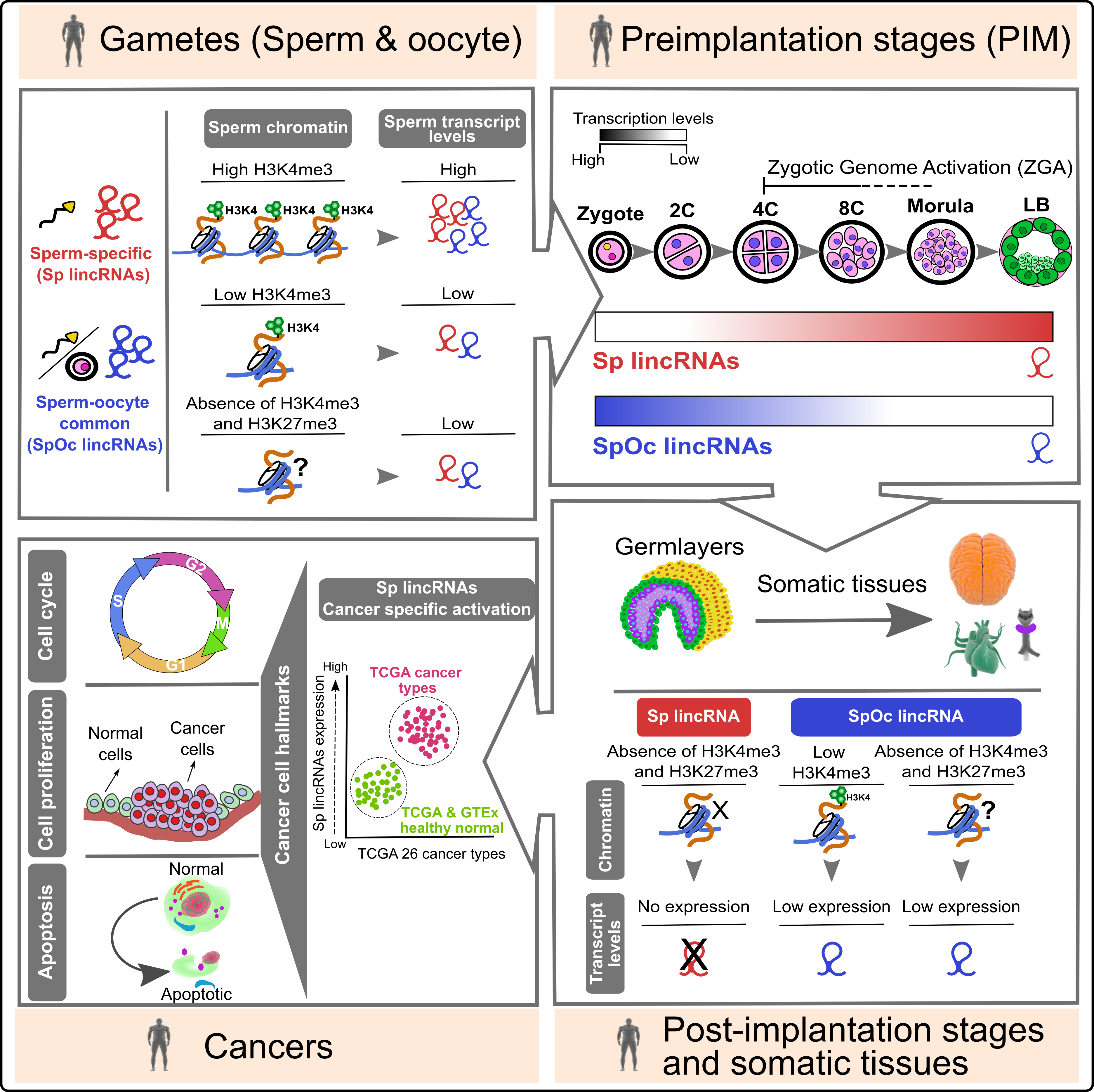 |
Sperm barcoded chromatin imprints and lincRNAs: For long, sperm considered a passive vehicle, mere transmitting the paternal genome into oocyte during fertilization. Contrary to the prevailing view on sperm, recent studies have shown that sperm chromatin can influence gene expression during preimplantation development. However, the importance of sperm encoded transcripts and chromatin imprints in organismal development is poorly investigated. In this study, we show that human sperm and oocyte carry equal number of transcripts. Sperm and sperm-oocyte transcripts carry distinct chromatin structures, correlating with their transcript levels. Sperm-specific transcripts are not expressed in early preimplantation embryos but become activated during the onset of zygotic genome activation, whereas sperm-oocyte expressed transcripts remain active during early preimplantation development but starts to degrade at the onset of zygotic genome activation. |
|
Interestingly, sperm-specific lincRNA promoters carry distinct chromatin structures that are exclusive to sperm and not present in the embryonic germ layers and somatic tissues. Additionally, and more importantly, sperm-specific lincRNAs showed cancer-specific activation. |
|